Difference between revisions of "Software for integrating ODEs"
Kevin Dunn (talk | contribs) m (→Source code) |
Kevin Dunn (talk | contribs) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 7: | Line 7: | ||
== Example == | == Example == | ||
Consider the pair of differential equations (covered in | Consider the pair of differential equations (covered in class on [[Isothermal_reactor_design_-_2013|11 February]]): | ||
<rst> | <rst> | ||
<rst-options: 'toc' = False/> | <rst-options: 'toc' = False/> | ||
| Line 32: | Line 32: | ||
But there are a few other unknowns we must first specify: | But there are a few other unknowns we must first specify: | ||
* :math:`r_A' = -k' \dfrac{(1-X)y}{(1+\varepsilon | * :math:`r_A' = -k' \dfrac{(1-X)y}{(1+\varepsilon X)}` | ||
* :math:`q = \dfrac{(1 + \varepsilon X)}{y}` | * :math:`q = \dfrac{(1 + \varepsilon X)}{y}` | ||
* :math:`F_{A0} = 0.1362\,\text{mol.s}^{-1}` | * :math:`F_{A0} = 0.1362\,\text{mol.s}^{-1}` | ||
| Line 39: | Line 39: | ||
* :math:`\varepsilon = -0.15` | * :math:`\varepsilon = -0.15` | ||
</rst> | </rst> | ||
== Polymath== | == Polymath== | ||
| Line 70: | Line 71: | ||
== Python == | == Python == | ||
=== Installing Python === | === Installing Python === | ||
Installation differs from Windows to Linux to Mac. One Python distribution that works well for many people is the [ | Installation differs from Windows to Linux to Mac. One Python distribution that works well for many people is the [https://www.enthought.com/ Enthought Python Distribution] (EPD) and the other is [https://www.continuum.io/downloads Anaconda]. Another version, especially for Windows users is [https://python-xy.github.io/ Python(x,y)]. | ||
=== Libraries required === | === Libraries required === | ||
| Line 236: | Line 237: | ||
% Integrate the ODE(s): | % Integrate the ODE(s): | ||
[ | [indep, depnt] = ode45(@pbr, [indep_start, indep_final], [X_depnt_zero, y_depnt_zero]); | ||
% Plot the results: | % Plot the results: | ||
clf; | clf; | ||
plot( | plot(indep, depnt(:,1), 'b') | ||
grid('on') | grid('on') | ||
hold('on') | hold('on') | ||
plot( | plot(indep, depnt(:,2), 'g') | ||
xlabel('Catalyst weight, W [kg]') | xlabel('Catalyst weight, W [kg]') | ||
ylabel('X and y') | ylabel('X and y') | ||
legend('X', 'y') | legend('X', 'y') | ||
</syntaxhighlight> | </syntaxhighlight> | ||
Latest revision as of 19:41, 6 January 2017
Background
Numerically integrating ODE's is something you should have mastered in your pre-requisite 3E4 course. If you are not familiar with this topic, it is your responsibility to quickly catch up, because we will use this intensively for the rest of the course.
Here is a tutorial a wrote a few years ago when I taught 3E4 in 2010. The tutorial below is similar, but uses a reactor design example.
Example
Consider the pair of differential equations (covered in class on 11 February): <rst> <rst-options: 'toc' = False/> <rst-options: 'reset-figures' = False/> .. math::
\dfrac{dX}{dW} &= \dfrac{-r_A'}{F_{A0}} \\ \dfrac{dy}{dW} &= -\dfrac{\alpha}{2y}\left(1 + \varepsilon X\right)
Some terminology (recap from your pre-requisite math courses)
* The independent variable is \(W\) * The two dependent variables (variables being integrated with respect to the independent variable) are \(X\) and \(y\)
Since there are two dependent variables, we require initial conditions for each variable. In this case:
* :math:`X(0) = 0.0` * :math:`y(0) = 1.0`
We also need to specify initial **and final conditions** for the independent variable:
* :math:`W` initially is 0.0 at the reactor entrance * :math:`W` finally is 28.0 kg at the reactor exit
But there are a few other unknowns we must first specify:
* :math:`r_A' = -k' \dfrac{(1-X)y}{(1+\varepsilon X)}` * :math:`q = \dfrac{(1 + \varepsilon X)}{y}` * :math:`F_{A0} = 0.1362\,\text{mol.s}^{-1}` * :math:`k' = 0.0074\,\text{mol.s}^{-1}\text{.(kg catalyst)}^{-1}` * :math:`\alpha = 0.0367\,\text{kg}^{-1}` * :math:`\varepsilon = -0.15` </rst>
Polymath
Download and install Polymath from the CD/DVD included with your course textbook (Windows only; sorry Mac and Linux versions are not available). Unfortunately this is only a 15 day version, which is pretty useless for this course, since you will require the code for at least the next 30 to 45 days. So rather use one of the other options below, if possible.
Enter the following Polymath code (as demonstrated in class on 11 February. Check the video out for a detailed explanation of the code.
# Differential equations
d(X) / d(W) = -rAdash / FA0
X(0) = 0
d(y) / d(W) = -alpha/(2*y) * (1+eps*X)
y(0) = 1.0
# Constants
FA0 = 0.1362 # [mol/s]
kdash = 0.0074 # [mol/(kg catalyst . s)]
alpha = 0.0367 # [1/kg]
eps = -0.15 # [-]
# Algebraic equations
rAdash = -kdash * (1-X)/(1+eps*X) * y
flow_ratio = (1 + eps*X)/y
# Initial and final values for independent variable:
W(0) = 0
W(f) = 28
Ensure that you obtain a graphical output as shown here. This represents the profile of conversion, \(X\) and the pressure drop ratio \(y\) throughout the reactor.
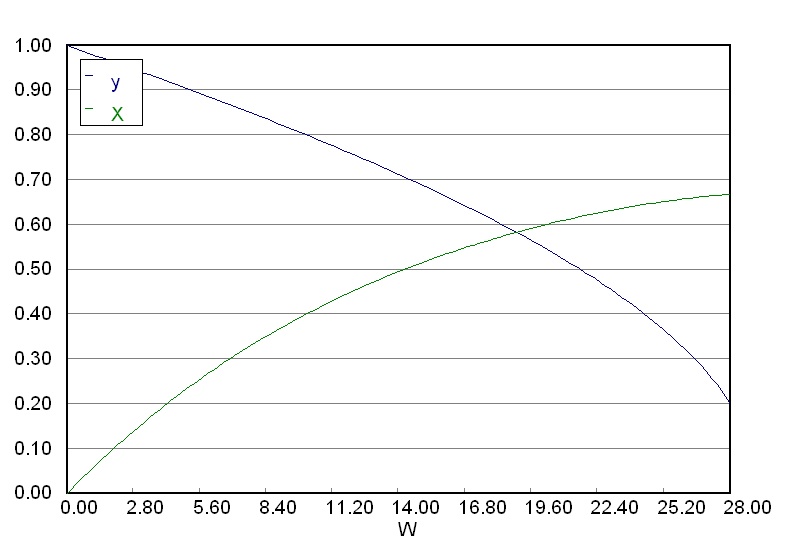
Python
Installing Python
Installation differs from Windows to Linux to Mac. One Python distribution that works well for many people is the Enthought Python Distribution (EPD) and the other is Anaconda. Another version, especially for Windows users is Python(x,y).
Libraries required
You are ready to go if you enter the following without getting error messages:
import numpy as np
from scipy import integrate
from matplotlib.pylab import (plot, grid, xlabel, ylabel)
If you experience problems importing these libraries, then re-install your Python distribution using one of the two options listed above.
Source code
import numpy as np
from scipy import integrate
from matplotlib.pylab import (plot, grid, xlabel, ylabel, show, legend)
def PBR_model(indep, depnt):
"""
Dynamic balance for the packed bed reactor (PBR); demo problem class 05C
indep: the independent ODE variable, such as time or length
depnt: a vector of dependent variables
X = depnt[0] = the conversion
y = depnt[1] = the pressure ratio = P/P_0 = y
Returns d(depnt)/d(indep) = a vector of ODEs
"""
# Assign some variables for convenience of notation
X = depnt[0]
y = depnt[1]
# Constants
FA0 = 0.1362 # [mol/s]
kdash = 0.0074 # [mol/(kg catalyst . s)]
alpha = 0.0367 # [1/kg]
eps = -0.15 # [-]
#Algebraic equations
rAdash = -kdash * (1-X)/(1+eps*X) * y
# Output from this ODE function must be a COLUMN vector, with n rows
n = len(depnt)
d_depnt__d_indep = np.zeros((n,1))
d_depnt__d_indep[0] = -rAdash / FA0
d_depnt__d_indep[1] = -alpha/(2*y) * (1+eps*X)
return d_depnt__d_indep
# The "driver" that will integrate the ODE(s):
# -----------
# Start by specifying the integrator:
# use ``vode`` with "backward differentiation formula"
r = integrate.ode(PBR_model).set_integrator('vode', method='bdf')
# Set the independent variable's range
indep_start = 0.0 # kg
indep_final = 28.0 # kg
delta = 1.0 # the results will be shown only at these ``delta`` points
num_steps = np.floor((indep_final - indep_start)/delta) + 1 # Number of steps: 1 extra for initial condition
# Set initial condition(s): for integrating variables (dependent variables)
X_depnt_zero = 0.0 # i.e. X(W=0) = 0.0
y_depnt_zero = 1.0 # i.e. y(W=0) = 1.0
r.set_initial_value([X_depnt_zero, y_depnt_zero], indep_start)
# Create vectors to store trajectories
W = np.zeros((num_steps, 1))
X = np.zeros((num_steps, 1))
y = np.zeros((num_steps, 1))
W[0] = indep_start
X[0] = X_depnt_zero
y[0] = y_depnt_zero
# Integrate the ODE(s) across each delta
k = 1
while r.successful() and k < num_steps:
r.integrate(r.t + delta)
# Store the results to plot later
W[k] = r.t
X[k] = r.y[0]
y[k] = r.y[1]
k += 1
# All done! Plot the trajectories:
plot(W, X)
grid('on')
xlabel('Catalyst weight, W [kg]')
plot(W, y)
ylabel('X and y [both dimensionless]')
legend(['X', 'y'])
show()
Ensure that you obtain a graphical output as shown here. This represents the profile of conversion, \(X\) and the pressure drop ratio \(y\) throughout the reactor.

MATLAB
With MATLAB we create two files: a file containing the model equation, and another that drives the ODE integrator. Once you have both files on your computer, call ODEdriver from the MATLAB command line.
Model file: pbr.m
function d_depnt__d_indep = pbr(indep, depnt)
% Dynamic balance for the packed bed reactor (PBR); demo problem class 05C
%
% indep: the independent ODE variable, such as time or length
% depnt: a vector of dependent variables
%
% X = depnt(1) = the conversion
% y = depnt(2) = the pressure ratio = P/P_0 = y
%
% Returns d(depnt)/d(indep) = a vector of ODEs
% Assign some variables for convenience of notation
X = depnt(1);
y = depnt(2);
% Constant(s)
FA0 = 0.1362; % [mol/s]
kdash = 0.0074; % [mol/(kg catalyst . s)]
alpha = 0.0367; % [1/kg]
eps = -0.15; % [-]
% Algebraic equation(s)
rAdash = -kdash * (1-X)/(1+eps*X) * y;
% Output from this ODE function must be a COLUMN vector, with n rows
n = numel(depnt);
d_depnt__d_indep = zeros(n,1);
d_depnt__d_indep(1) = -rAdash / FA0;
d_depnt__d_indep(2) = -alpha/(2*y) * (1+eps*X);
ODE integrator: ODEdriver.m
% Integrate the ODE
% -----------------
% The independent variable always requires an initial and final value:
indep_start = 0.0; % kg
indep_final = 28.0; % kg
% Set initial condition(s): for integrating variables (dependent variables)
X_depnt_zero = 0.0; % i.e. X(W=0) = 0.0
y_depnt_zero = 1.0; % i.e. y(W=0) = 1.0
% Integrate the ODE(s):
[indep, depnt] = ode45(@pbr, [indep_start, indep_final], [X_depnt_zero, y_depnt_zero]);
% Plot the results:
clf;
plot(indep, depnt(:,1), 'b')
grid('on')
hold('on')
plot(indep, depnt(:,2), 'g')
xlabel('Catalyst weight, W [kg]')
ylabel('X and y')
legend('X', 'y')